Most COVID-19 vaccine candidates in development worldwide were modelled on the original strain of the SARS-CoV-2 virus, the "D-strain", which was common amongst sequences published early in the pandemic.
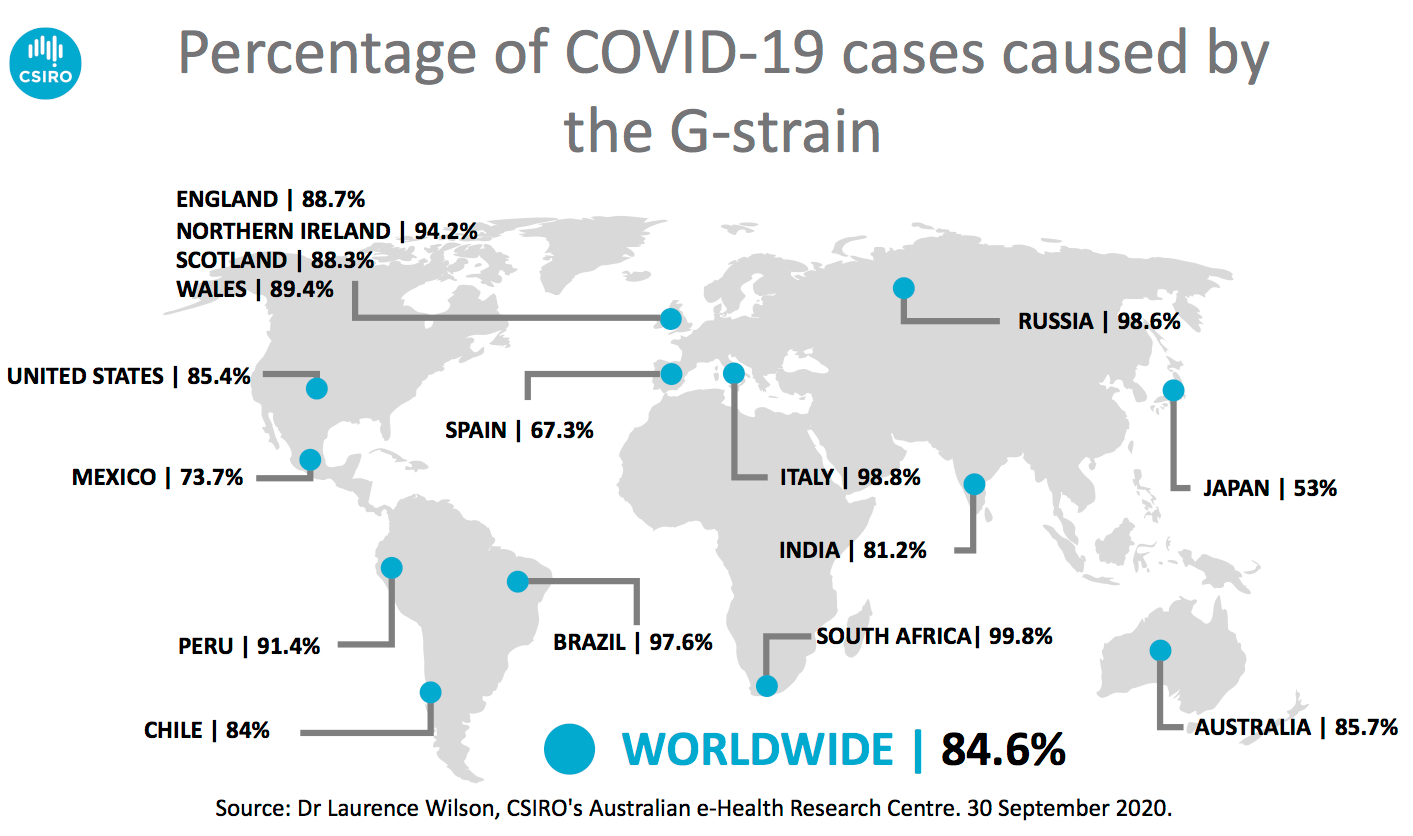
Analyses of published virus genomes showed the next most dominant strain globally was the "G-strain", which accounted for about 85 per cent of published SARS-CoV-2 genomes.
There had been fears that the G-strain, or "D614G" mutation within the main protein on the surface of the virus, could negatively impact vaccines under development.
At our Australian Centre for Disease Preparedness, our Dangerous Pathogens Team tested blood samples from ferrets vaccinated with Inovio Pharmaceuticals' INO-4800 candidate against virus strains that either possessed or lacked this D614G mutation.
Researchers found no evidence that the change would adversely impact the efficacy of the vaccine candidate. These results were published in npj Vaccines on 8 October 2020.
The results of the study were further interpreted with biomolecular modelling conducted by CSIRO's Data61, enabling interactions between the vaccine and virus to be simulated and visualised.
Most COVID-19 vaccine candidates in development worldwide were modelled on the original strain of the SARS-CoV-2 virus, the "D-strain", which was common amongst sequences published early in the pandemic.
Map of the world showing the percentage of COVID-19 cases caused by the G-strain. Percentage per country listed includes: Source: Dr Laurence Wilson, CSIRO's Australian e-Health Research Centre. 30 September 2020.
Analyses of published virus genomes showed the next most dominant strain globally was the "G-strain", which accounted for about 85 per cent of published SARS-CoV-2 genomes.
There had been fears that the G-strain, or "D614G" mutation within the main protein on the surface of the virus, could negatively impact vaccines under development.
At our Australian Centre for Disease Preparedness, our Dangerous Pathogens Team tested blood samples from ferrets vaccinated with Inovio Pharmaceuticals' INO-4800 candidate against virus strains that either possessed or lacked this D614G mutation.
Researchers found no evidence that the change would adversely impact the efficacy of the vaccine candidate. These results were published in npj Vaccines on 8 October 2020.
The results of the study were further interpreted with biomolecular modelling conducted by CSIRO's Data61, enabling interactions between the vaccine and virus to be simulated and visualised.
