We used bioinformatics algorithms to analyse the genome of the virus that causes COVID-19, to understand how changes in the virus affect its behaviour and impact.
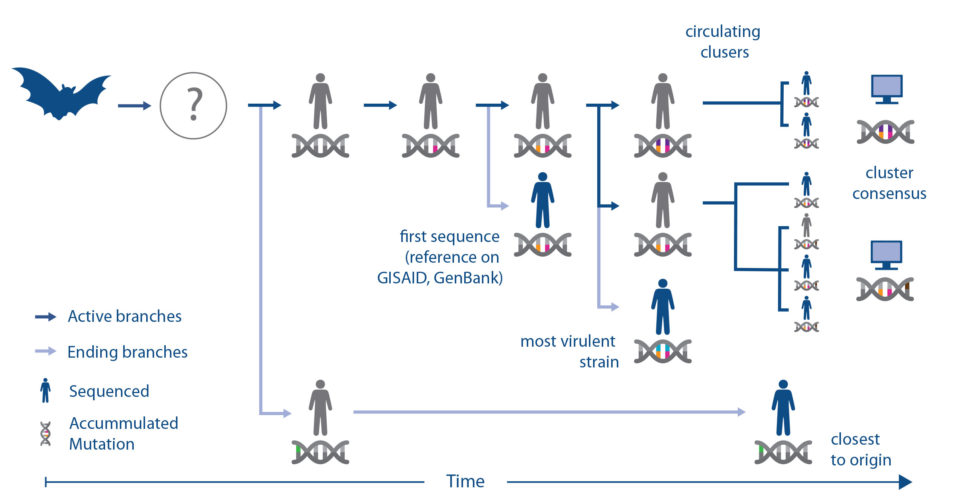 Coronaviruses have RNA instead of DNA, and RNA can change or mutate rapidly. These changes may need to be taken into account when testing which vaccines or treatments might work. However, identifying the mutations – or differences – among the 30,000 letters of the viral genome is not an easy task, akin to finding a needle in a haystack.
Coronaviruses have RNA instead of DNA, and RNA can change or mutate rapidly. These changes may need to be taken into account when testing which vaccines or treatments might work. However, identifying the mutations – or differences – among the 30,000 letters of the viral genome is not an easy task, akin to finding a needle in a haystack.
Researchers from our Australian e-Health Research Centre, Australian Centre for Disease Preparedness, and Health and Biosecurity teams adapted bioinformatics algorithms originally used to analyse the human genome, and used them to pinpoint the differences among thousands of different genetic sequences of the virus that causes COVID-19.
In a paper published by the Transboundary and Emerging Diseases journal on 19 April 2020, Supporting pandemic response using genomics and bioinformatics: a case study on the emergent SARS-CoV-2 outbreak, our researchers analysed the first 181 published genome sequences from the COVID-19 pandemic. The team confirmed that the virus was evolving into distinct clusters in different parts of the world.
They also called on the international community to share more genomic sequences of the virus alongside de-identified information about clinical symptoms and co-morbidities, in order to monitor the changes and form a better understanding of how important genetic differences are to the disease’s progression.
The COVID-19 genome visualisation platform can be found at Genomic Signature analysis of SARS-CoV-2.
Learn more: Genomics joins the fight against COVID-19
We used bioinformatics algorithms to analyse the genome of the virus that causes COVID-19, to understand how changes in the virus affect its behaviour and impact.
Coronaviruses have RNA instead of DNA, and RNA can change or mutate rapidly. These changes may need to be taken into account when testing which vaccines or treatments might work. However, identifying the mutations – or differences – among the 30,000 letters of the viral genome is not an easy task, akin to finding a needle in a haystack.
Researchers from our Australian e-Health Research Centre, Australian Centre for Disease Preparedness, and Health and Biosecurity teams adapted bioinformatics algorithms originally used to analyse the human genome, and used them to pinpoint the differences among thousands of different genetic sequences of the virus that causes COVID-19.
In a paper published by the Transboundary and Emerging Diseases journal on 19 April 2020, Supporting pandemic response using genomics and bioinformatics: a case study on the emergent SARS-CoV-2 outbreak, our researchers analysed the first 181 published genome sequences from the COVID-19 pandemic. The team confirmed that the virus was evolving into distinct clusters in different parts of the world.
They also called on the international community to share more genomic sequences of the virus alongside de-identified information about clinical symptoms and co-morbidities, in order to monitor the changes and form a better understanding of how important genetic differences are to the disease’s progression.
The COVID-19 genome visualisation platform can be found at Genomic Signature analysis of SARS-CoV-2.
